Aprendizado de Máquina
Dicas práticas
Prof. Ronaldo Cristiano Prati
Bloco A, sala 513-2
Dicas de SKLearn
- SKlearn é um pacote python muito que implementa vários algoritmos de aprendizado de máquina
- Ele também oforece um conjunto de funções que implementam diversas funcionalidades para utilizar esses algoritmos
Dicas de SKLearn
-
Vamos ver como criar a curva de aprendizado (incrementando o conjunto de dados) e a curva de validação (alteranando os valores do parâmetro livre)
-
Pode ser usado para analisar o trade-off bias-variância.
Pacotes
import matplotlib.pyplot as plt # data sets from sklearn.datasets import load_digits, load_iris # Algoritmos from sklearn.linear_model import LogisticRegression from sklearn.svm import SVC from sklearn.model_selection import learning_curve, validation_curve import numpy as np
Curva de Aprendizado
# Seleciona 2 classes do dataset digits X,y = load_digits(n_class=2,return_X_y=True) # cria uma lista de 0.1 a 1, contendo 10 pontos igualmente espaçados sizes = np.linspace(0.1,1,10) algs = { 'Logistic Regression' : LogisticRegression(solver='lbfgs'), 'SVM - Gaussian' : SVC()} for name, alg in algs.items(): train_sizes, train_scores, test_scores = learning_curve( alg, X, y, cv=3, train_sizes=sizes) plot_learning_curve(train_sizes, train_scores, test_scores,name)
Curva de Aprendizado
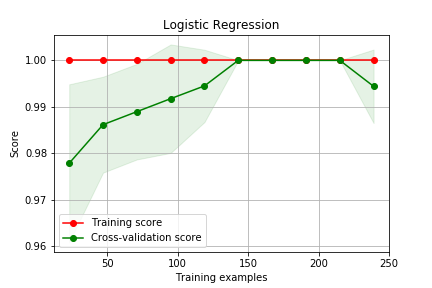
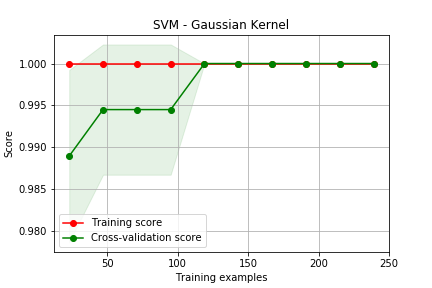
Curva de Validação
X,y = load_digits(return_X_y=True) # cria pontos exponencialmente expaçados 10^-6 até 10^-1 param_range = np.logspace(-6, -1, 5) train_scores, test_scores = validation_curve( SVC(), X, y, param_name="gamma", param_range=param_range, cv=5, scoring="accuracy", n_jobs=1)
Curva de Validação
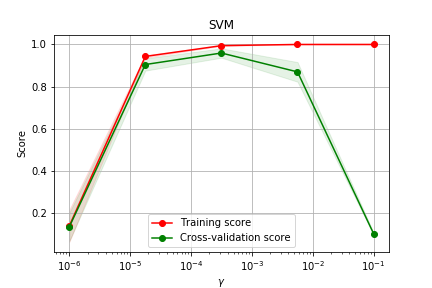
Grid Search
-
Em algumas situações queremos variar mais de um parâmetro livre do algoritmo.
- Em SVMs por exemplo, podemos querer ajustar o parâmetro de regularização C e o parâmetro do kernel σ
-
GridSearch faz uma busca combinando todas as possibilidades para esses valores
Grid Search
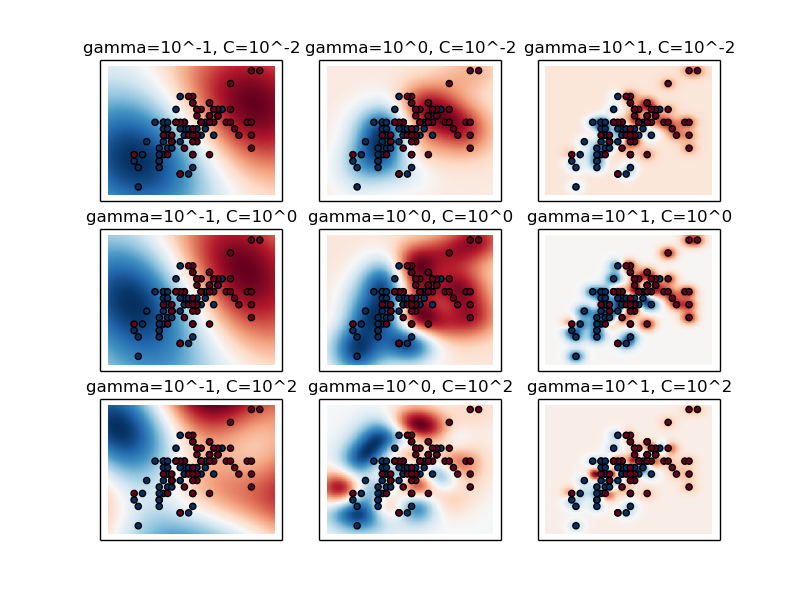
X, y = load_iris(return_X_y=True) # faixa de valores de C C_range = np.logspace(-2, 10, 13) # faixa de valores de gamma gamma_range = np.logspace(-9, 3, 13) param_grid = dict(gamma=gamma_range, C=C_range) cv = StratifiedShuffleSplit(y, n_iter=5, test_size=0.2) grid = GridSearchCV(SVC(), param_grid=param_grid, cv=cv) grid.fit(X, y)
Grid Search
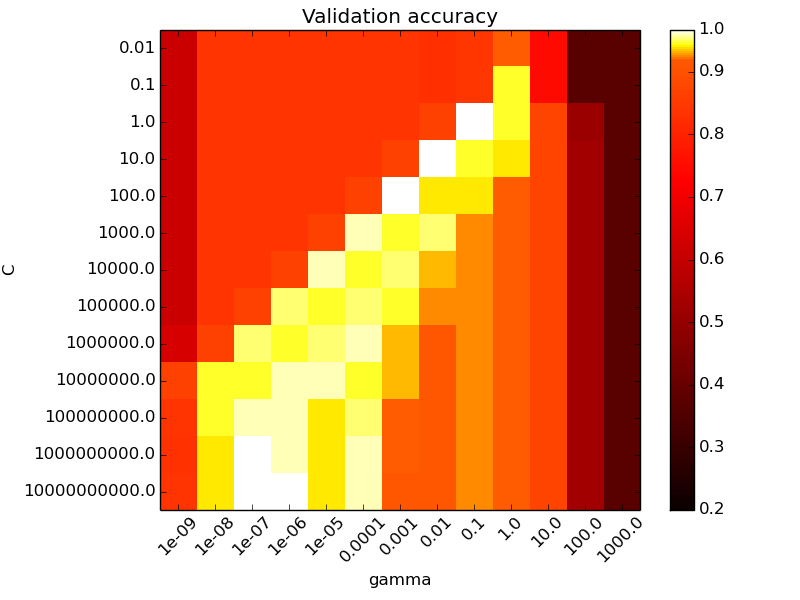
The best parameters are {'C': 1.0, 'gamma': 0.1} with a score of 0.97
Grid Search
Validação cruzada
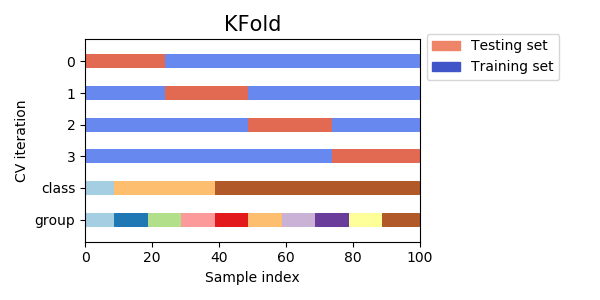
- Como visto anteriormente, em validação cruzada, dividimos o conjunto em diferentes partes de treino e teste
- Se a base estiver de alguma maneira ordenada, temos que tomar cuidado em como fazemos a divisão
Validação cruzada
- O Sklearn tem vários metodos que auxiliam a fazer validação cruzada.
- O mais simples deles divide os as linhas da base em blocos igualmente espaçados.
Validação cruzada
from sklearn.model_selection import KFold from sklearn.metrics import accuracy_score X, y = load_iris(return_X_y=True) alg = LogisticRegression(solver='lbfgs',multi_class='ovr') accuracies = [] kf = KFold(n_splits=5) for train, test in kf.split(X): X_train = X[train] X_test = X[test] y_train = y[train] y_test = y[test] alg.fit(X_train,y_train) pred = alg.predict(X_test) accuracies.append(accuracy_score(y_test,pred))
Validação cruzada
print(accuracies) [1.0, 0.9, 0.5, 0.9333333333333333, 0.6333333333333333] print (np.mean(accuracies),np.std(accuracies)) 0.7933333333333332 0.19252705437591536
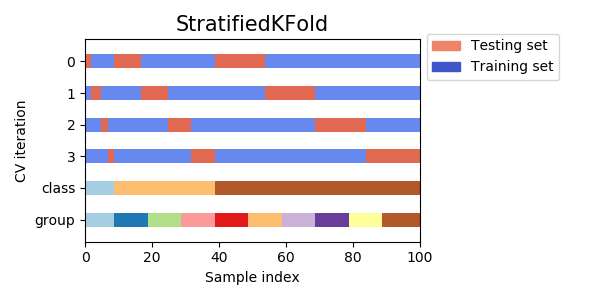
Validação cruzada estratificada
- Esse grande variação vem do fato que, ao fazermos a validação cruzada, pegamos blocos contíguos
- Como a base está agrupada por classes, temos conjuntos com classes não representadas no treino/teste
- Validação cruzada estratificada leva em consideração as classes para fazer a divisão
Validação cruzada estratificada
from sklearn.model_selection import StratifiedKFold from sklearn.metrics import accuracy_score X, y = load_iris(return_X_y=True) alg = LogisticRegression(solver='lbfgs',multi_class='ovr') accuracies = [] kf = StratifiedKFold(n_splits=5) for train, test in kf.split(X,y): X_train = X[train] X_test = X[test] y_train = y[train] y_test = y[test] alg.fit(X_train,y_train) pred = alg.predict(X_test) accuracies.append(accuracy_score(y_test,pred))
Validação cruzada estratificada
print(accuracies) [0.8666666666666667, 0.9666666666666667, 0.9333333333333333, 0.9333333333333333, 1.0] print (np.mean(accuracies),np.std(accuracies)) 0.9400000000000001 0.044221663871405324
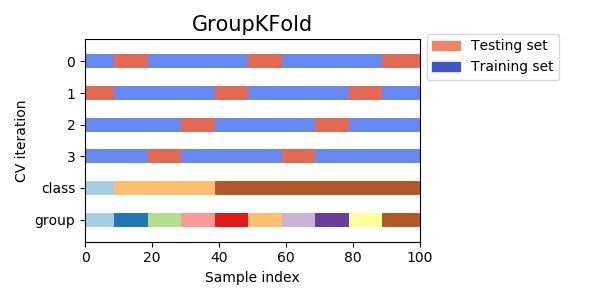
Validação cruzada agrupada
- Em algumas situações, a base de dados também pode estar organizada em grupos
- Por exemplo, uma prática comum em classificação de imagens, por exemplo, é rotacionar a imagem em 90o,180o,270o e 360o
- Nesse caso, para cada imagem inicial, temos outras 4
- Para esses casos, devemos usar validação cruzada agrupada
Validação cruzada agrupada
from sklearn.model_selection import GroupKFold X = [0.1, 0.2, 2.2, 2.4, 2.3, 4.55, 5.8, 8.8, 9, 10] y = ["a", "b", "b", "b", "c", "c", "c", "d", "d", "d"] groups = [1, 1, 1, 2, 2, 2, 3, 3, 3, 3] gkf = GroupKFold(n_splits=3) for train, test in gkf.split(X, y, groups=groups): print("%s %s" % (train, test)) [0 1 2 3 4 5] [6 7 8 9] [0 1 2 6 7 8 9] [3 4 5] [3 4 5 6 7 8 9] [0 1 2]
Validação cruzada agrupada
Métrica de avaliação
- Em diversas aplicações, a escolha da métrica de avaliação pode levar a resultados incosistentes
- Por exemplo, quando temos classes desbalanceadas, é fácil ter uma alta acurácia
- Vamos assumir que fraude de cartão só ocorre 0.1% das transações
- Classificador que nunca preve fraude acerta 99.9% das vezes!
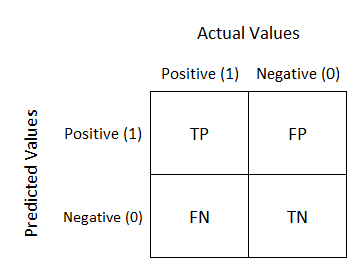
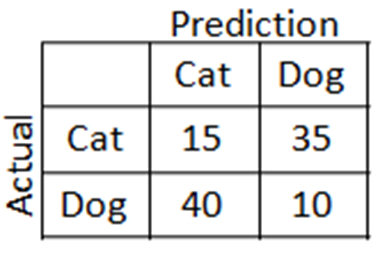
Matriz de confusão
Precisão (precision)
-
Avalia cada classe individualmente
-
Vamos chamar a nossa classe de interesse positiva
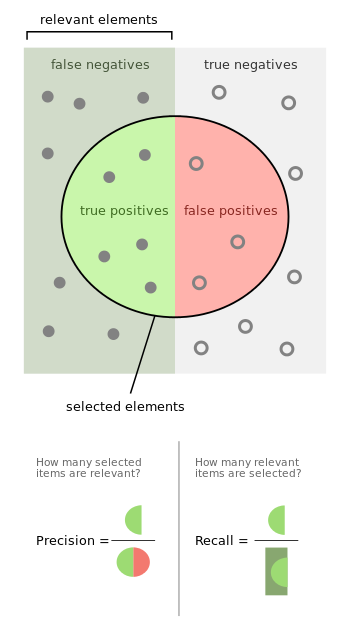
- Quando o algoritmo prediz um exemplo como positivo, com que frequência ele acerta aquela predição?
precision=TP+FPTP
Revocação (recall)
-
Avalia cada classe individualmente
-
Vamos chamar a nossa classe de interesse positiva
- Com relação a todos os exemplos positivos do conjunto de teste, quantos o algoritmo acertou?
recall=TP+FNTP
Métrica de avaliação
- Geralmente escolhemos a classe minoritária como a de interesse
- Doença rara, fraude, etc.
- Calcular precisão e revocação para essa classe nos dá uma ideia melhor do desempenho do algoritmo para aquela classe
- Eventualmente podemos computar para outra(s) classe(s) também
F1-measure
- A medida F1 é muito usada em algumas áreas como um compromisso entre precisão e revocação.
- Média harmônica entre precisão e revocação
F1=2⋅precision+recallprecision∗recall
Trade-off entre precisão e revocação
- Para muitas aplicações, queremos controlar o trade-off entre precisão e revocação
- É fácil otimizar um ou outro:
- precisão: só predizar o mais seguro (e.g., o de maior probabilidade)
- revocação: predizer todos os exemplos como positivos
- Entretanto, otimizar os dois pode ser difícil
Trade-off entre precisão e revocação
- Uma maneira de controlar o trade-off entre essas duas medidas é variar o limiar da fronteira de decisão
- Por exemplo, na regressão logística, o algoritmo dá como saída a probabilidade da classe
Trade-off entre precisão e revocação
- Normalmente assumimos que a fronteira está em predizer classe positiva se hθ(x)>0.5.
- Por exemplo, podemos aumentar precisão predizendo positivo se hθ(x)>0.8;
- ou aumentar a revocação predizendo positivo se hθ(x)>0.2
Trade-off entre precisão e revocação
- A escolha do limiar é dependente do problema
- Podemos analisar o desempenho de um modelo considerando todos os possíves limiares
- Curva precisão-revocação (precision-recall)
- Precision (eixo y)
- Recall (eixo x)
- Para cada possível limiar, calcula-se um valor de precision e recall
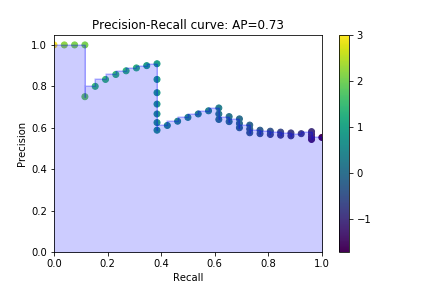
Precison-recall curve
from sklearn.metrics import precision_recall_curve from sklearn.model_selection import train_test_split from sklearn.datasets import make_classification from sklearn import svm X,y = make_classification(class_sep=0.5) X_train, X_test, y_train, y_test = train_test_split(X, y,test_size=.5) alg = svm.LinearSVC() alg.fit(X_train, y_train) y_score = alg.decision_function(X_test) precision, recall, t = precision_recall_curve(y_test, y_score)
Precision recall curve